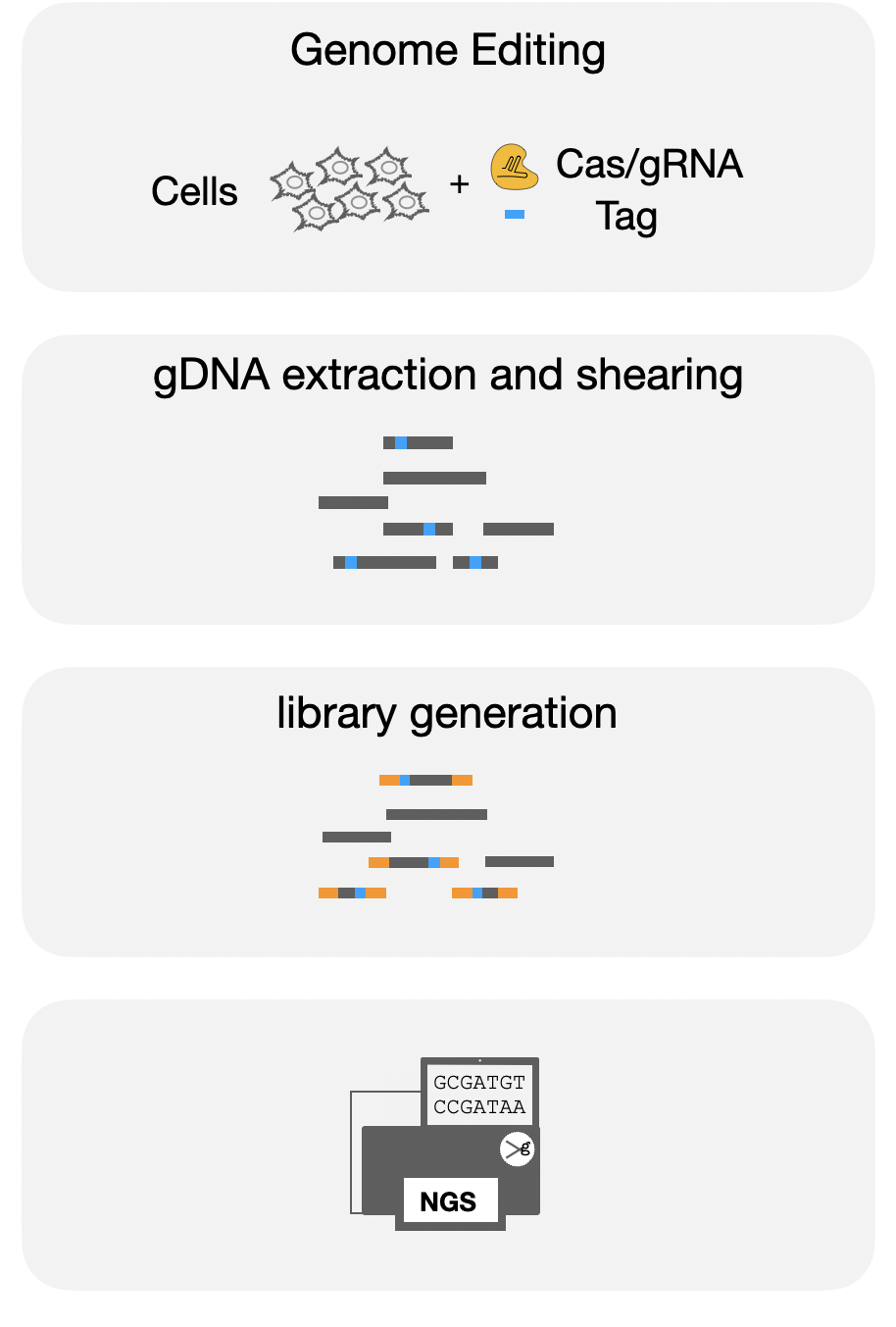
Off Target Identification
Off target identification is becoming increasingly important to create reliable tools for basic and translational research. The powerful NGS platforms allow us to identify erroneously targeted sites and accurately assess their prevalence.
Several approaches with different strengths and limitations have been described in the literature. In GEML, we offer three workflows for identifying off targets: iGUIDE/GUIDE-seq, CHANGE-seq, and DISCOVER-seq.
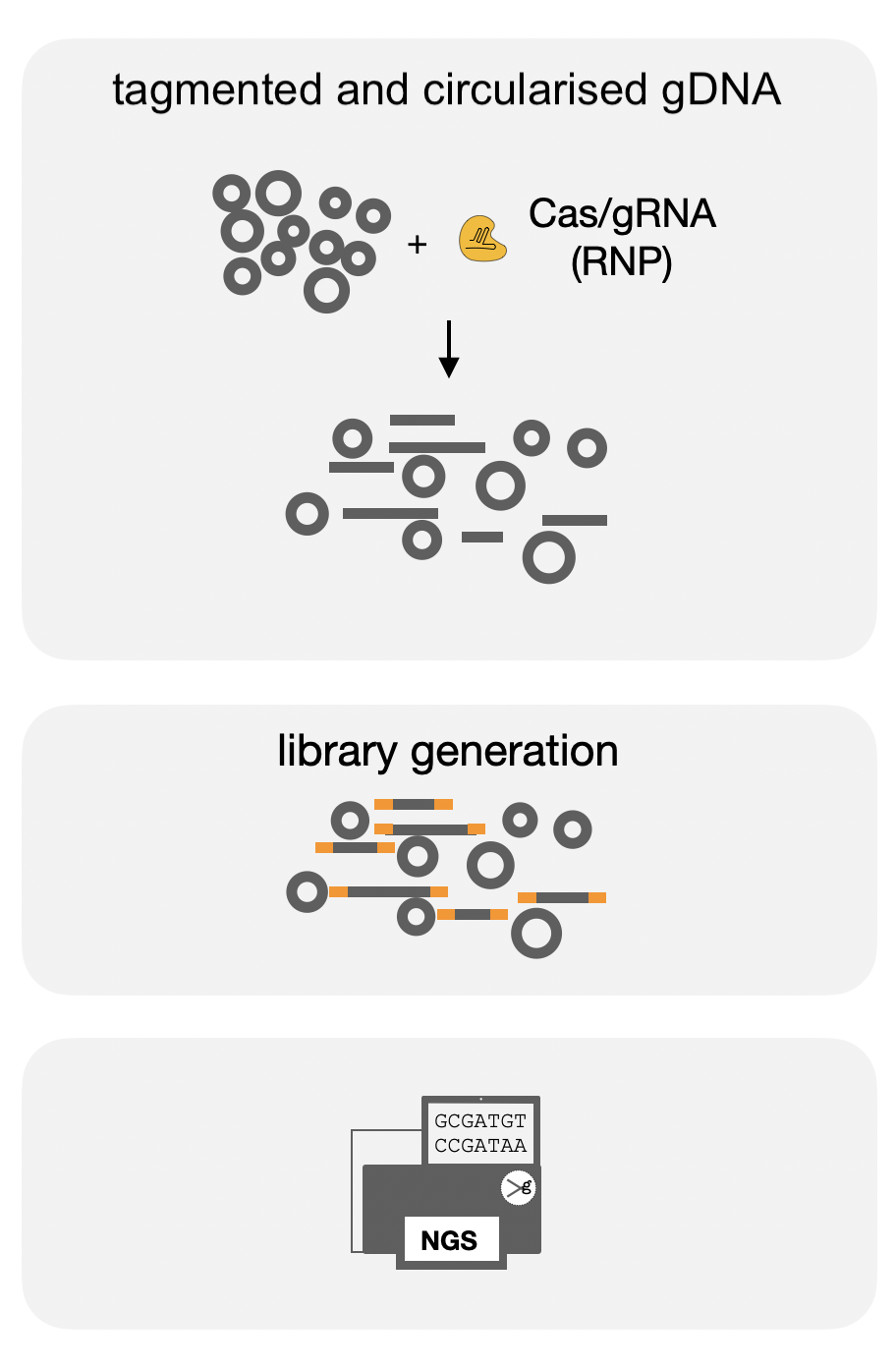
CHANGE-seq assesses the in vitro activity of guided nucleases. A circular library preparation of the genome of interest is required. Sites that are cut by the nuclease will contribute to a population of linear genomic fragments that will be used for NGS library preparation and sequencing.
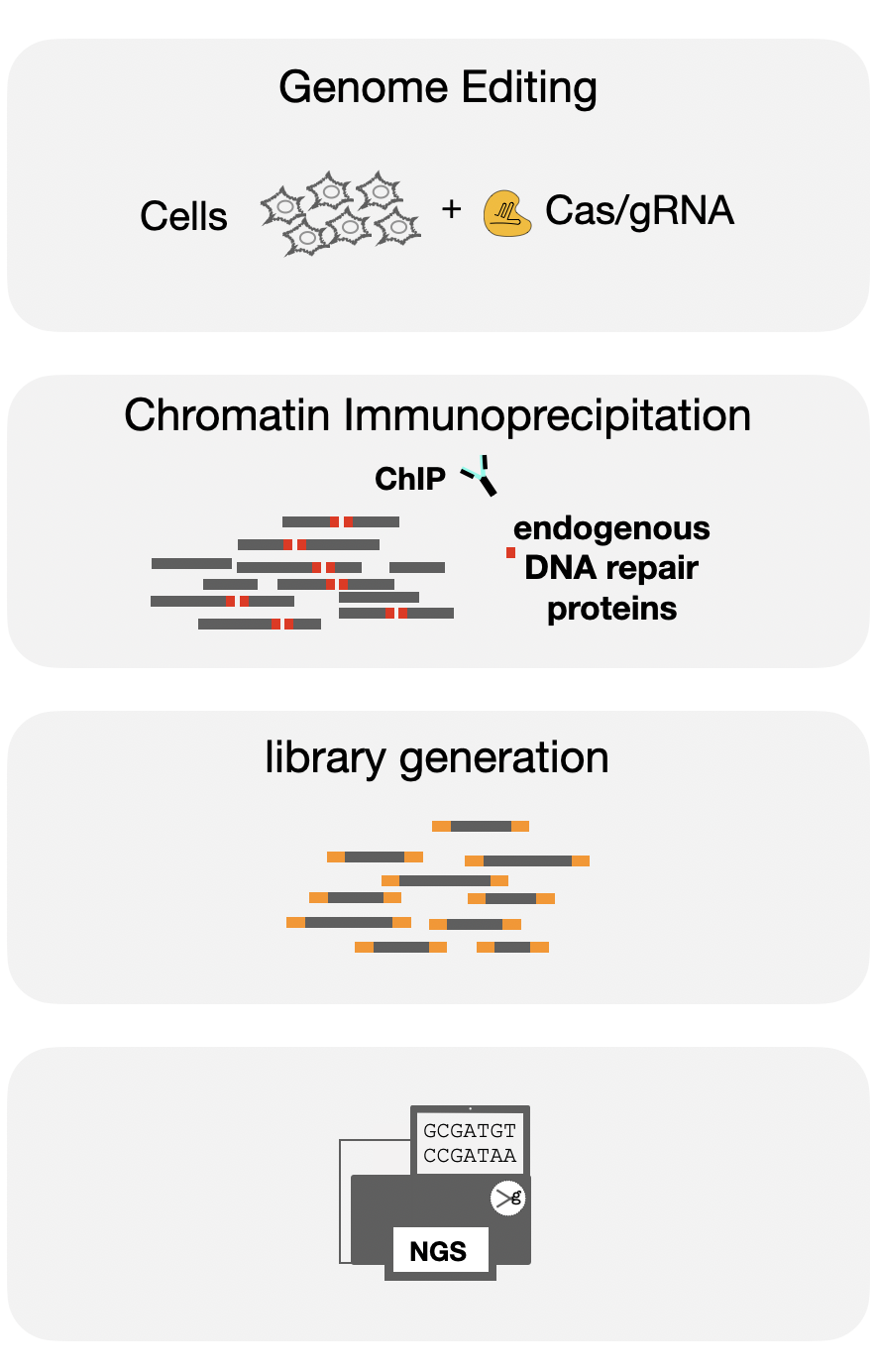
DISCOVER-seq takes advantage of the cell repair mechanisms to capture double stranded breaks as they appear. Using Chromatin immunoprecipitation of DNA repair proteins (e.g. MRE11) we can generate a library of sites that correspond to off-target activity of the genome engineering tool.
References:
external page Tsai SQ, Zheng Z et al Nat Biotechnol.2015 (GUIDE-Seq)
external page Nobles CL et al Genome Biology.2019 (iGUIDE)
external page Lazzarotto CR et al Nat Biotechnol.2020 (CIRCLE-Seq)
external page Wienert B, Wyman SK et al Science.2019 (DISCOVER-Seq)